[Top][All Lists]
[Date Prev][Date Next][Thread Prev][Thread Next][Date Index][Thread Index]
[Gnumed-devel] Incoming HL7 result microbiology report handling
|
From: |
Jim Busser |
|
Subject: |
[Gnumed-devel] Incoming HL7 result microbiology report handling |
|
Date: |
Wed, 06 Jul 2011 14:27:40 -0700 |
In the message below, 4th ORC OBR (Stool culture) we see a variety of OBX
patterns. The cleanest end result would be 4 rows in test_result:
- a val_alpha comment
Specimen #1\.br\
- 10 day cold enrichment for Yersinia \.br\
is performed on all stool specimens.\.br\
Further report will follow if positive.\.br
- Result phoned to physician's office \.br\
- a second val_alpha comment
- a third val_alpha row (Organism 1 result including *its* antibiotic
sensitivity information)
- a fourth val_alpha row (Organism 2 result, including all *its* antibiotic
sensitivity information)
Question:
If we stuff all the above Organism identification and culture and sensitivity
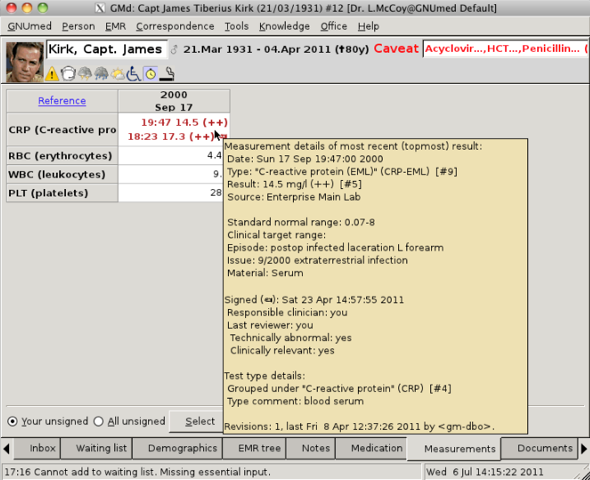
results in a test_result table cell, how does the user properly "see" it? A
tooltip comes to mind, *but* we already use measurement result tooltips to
display metadata (see screen shot).
-- Jim (sig 1)
*********************************************************************
Administrivia:
just my notes on what the program logic should have to identify. The message
contains…
- a few OBX segments which have a value type (field 3) of 'FT' which identify
- technique for Yersinia included
- result phoned
- organism 1 E coli light growth
- organism 2 Salmonella
- many more that are of value type 'ST' (short text)
The trick here is that the two OBX which are simply comments contain (to the
right of their LOINC^Lab name for test) an empty field for
OBX 005 Observation sub-ID
whereas the organisms (and the OBX pertaining to that organism) contain, in the
case of organism 1, a data type of "short text" with value of '1' and -- in the
case of organism 2 -- similar with sub-ID of '2'.
This means that when writing incoming message results, the relationship between
OBX and rows to be created in clin.test_result should be
where OBX 005 is empty ('||')
--> create a row and put result in val_alpha
where OBX 005 exhibits a change and is non-empty ('1')
--> create a row and put result in val_alpha
where OBX 005 exhibits no change and is non-empty (subsequent '1' s)
--> append to val_alpha:
(I would suggest an internal new line delimiter then ||) \
OBX 003 Observation Identifier (LOINC^lab result name) ||
'space' || \
OBX 005 'ST value' (here 'R / S' for Resistant / Sensitive)
-- Jim (sig 2)
******************************************
MSH|^~\&|SENDER_APP|MDS|HTTPCLIENT|vendor1|20050628140722||ORU^R01|MDC20050628140721957|P|2.3|||ER|AL
PID|||||DEMO^APATIENT||19770726|M|||260-4400 DOMI
AVENUE\.br\COQUITLAM\.br\BC\.br\||(604)999-9999
ORC|RE||05-994065004-SC-1|||||||||90909^MDCARE^BOB
OBR|5||05-994065004-SC-1|SC^Stool
Culture||20050506135300|20050506135300|||||||20050506135300||90909^MDCARE^BOB||||||20050512094510||MICRO|C|||90909^MDCARE^BOB
OBX|1|FT|X805^Comment||- Specimen #1\.br\- 10 day cold enrichment for Yersinia
\.br\is performed on all stool specimens.\.br\Further report will follow if
positive.\.br\||||||F|||20050512094510
OBX|2|FT|X10025^Comment||- Result phoned to physician's office
\.br\||||||F|||20050512094510
OBX|3|FT|X433^Organism 1|1|ESCHERICHIA COLI\.Zt\LIGHT
GROWTH\.br\||||||F|||20050512094510
OBX|4|ST|28-1^Ampicillin|1|S||||||F|||20050512094510
OBX|5|ST|X10042^Cephalothin/Cephalexin|1|R||||||C|||20050512094510
NTE|||Org1 Previously Reported As: S
OBX|6|ST|476-2^Sulfonamide|1|S||||||F|||20050512094510
OBX|7|ST|516-5^Trimethoprim-Sulfa|1|S||||||F|||20050512094510
OBX|8|ST|185-9^Ciprofloxacin|1|S||||||F|||20050512094510
OBX|9|ST|367-3^Norfloxacin|1|S||||||F|||20050512094510
OBX|10|ST|267-5^Gentamicin|1|S||||||F|||20050512094510
OBX|11|ST|496-0^Tetracycline|1|S||||||F|||20050512094510
OBX|12|ST|173-5^Chloramphenicol|1|S||||||F|||20050512094510
OBX|13|ST|351-7^Nalidixic Acid|1|S||||||F|||20050512094510
OBX|14|ST|363-2^Nitrofurantoin|1|S||||||F|||20050512094510
OBX|15|FT|X10019^Organism 2|2|SALMONELLA SPECIES\.Zt\MODERATE
GROWTH\.br\||||||F|||20050512094510
OBX|16|ST|28-1^Ampicillin|2|S||||||F|||20050512094510
OBX|17|ST|X10042^Cephalothin/Cephalexin|2|S||||||F|||20050512094510
OBX|18|ST|233-7^Erythromycin|2|S||||||F|||20050512094510
OBX|19|ST|516-5^Trimethoprim-Sulfa|2|S||||||F|||20050512094510
OBX|20|ST|185-9^Ciprofloxacin|2|S||||||F|||20050512094510
OBX|21|ST|367-3^Norfloxacin|2|S||||||F|||20050512094510
OBX|22|ST|267-5^Gentamicin|2|S||||||F|||20050512094510
OBX|23|ST|496-0^Tetracycline|2|S||||||F|||20050512094510
OBX|24|ST|173-5^Chloramphenicol|2|S||||||F|||20050512094510
OBX|25|ST|351-7^Nalidixic Acid|2|S||||||F|||20050512094510
OBX|26|ST|363-2^Nitrofurantoin|2|S||||||F|||20050512094510
OBX|27|ST|X084^Polymixin B|2|S||||||F|||20050512094510
-- Jim (sig 3)
- [Gnumed-devel] Incoming HL7 result microbiology report handling,
Jim Busser <=
- Re: [Gnumed-devel] How to display your results, richard terry, 2011/07/06
- Re: [Gnumed-devel] How to display your results, Jim Busser, 2011/07/06
- Re: [Gnumed-devel] How to display your results, Jim Busser, 2011/07/06
- Re: [Gnumed-devel] How to display your results, Karsten Hilbert, 2011/07/07
- Re: [Gnumed-devel] How to display your results, Jim Busser, 2011/07/07
- Re: [Gnumed-devel] How to display your results, Karsten Hilbert, 2011/07/07
- Re: [Gnumed-devel] How to display your results, Jim Busser, 2011/07/07
- Re: [Gnumed-devel] How to display your results, Karsten Hilbert, 2011/07/07
- Re: [Gnumed-devel] How to display your results, Karsten Hilbert, 2011/07/07
- Re: [Gnumed-devel] How to display your results, Karsten Hilbert, 2011/07/07
